Distributed mode (daal4py, CPU)
Introduction
Module daal4py within the Extension for Scikit-learn* offers distributed versions of some algorithms that can run on compute clusters managed through the MPI framework, optionally aided by mpi4py.
Compared to the SMPD mode in the sklearnex module which runs on multiple
GPUs, the distributed mode of algorithms in the daal4py module runs on CPU-based nodes - i.e.
it can be used to execute algorithms on multiple machines that hold different pieces of the data
each, communicating between themselves through the MPI framework; thereby allowing to scale the
same algorithms to much larger problem sizes.
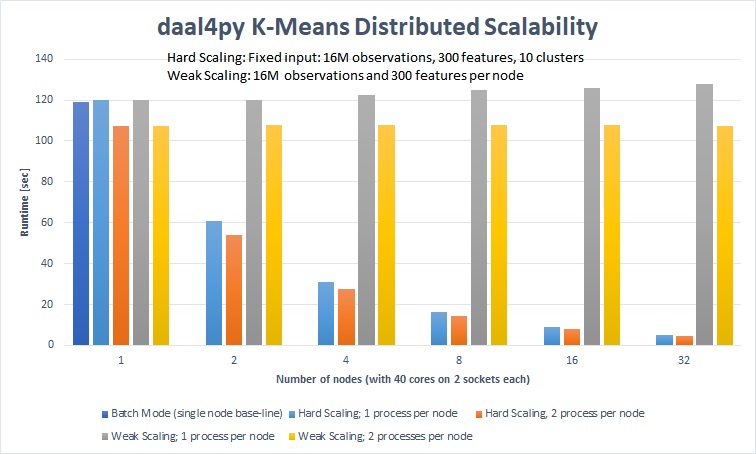
On a 32-node cluster (1280 cores) daal4py computed K-Means (10 clusters) of 1.12 TB of data in 107.4 seconds and 35.76 GB of data in 4.8 seconds.
Warning
Just like SPMD mode in sklearnex, using distributed mode in daal4py requires
the MPI runtime library managing the computations to be the same MPI backend library
with which the Extension for Scikit-learn* library was compiled, or to be ABI compatible with it.
Distributions of the Extension for Scikit-learn* in PyPI and conda-forge are both compiled with Intel’s MPI
as MPI backend (offered as Python package impi_rt in both PyPI and conda):
conda install -c conda-forge impi_rt mpi=*=impi
Using distributed mode with non-MPICH-compatible backends such as OpenMPI requires compiling the library from source with that backend.
See the docs for SPMD mode for more details.
Warning
If using distributed mode with the mpi4py library, that library must also be compiled
with the same MPI backend as the Extension for Scikit-learn*, or with a compatible MPI backend. A version
of mpi4py compiled with Intel’s MPI backend can be easily installed as follows (see docs
for SPMD mode for more details):
conda install -c conda-forge mpi4py mpi=*=impi
conda install -c https://software.repos.intel.com/python/conda/ -c conda-forge --override-channels mpi4py mpi=*=impi
Warning
Packages from the Intel channel are meant to be compatible with dependencies from conda-forge, and might not work correctly in environments that have packages installed from the anaconda channel.
pip install --index-url https://software.repos.intel.com/python/pypi mpi4py impi_rt
Using distributed mode
In order to use distributed mode, the code to execute must be saved in a Python file,
and this Python file must be executed through an MPI runner (mpiexec / mpirun).
The MPI runner in turn is the software that handles aspects like which nodes to use,
inter-node communication, and so on. The same Python code in the file will be executed
on all nodes, so the Python code may contain some logic to load the right subset of the
data on each node, aided for example by the process rank assigned by MPI.
From the daal4py side, in order to use distributed mode, algorithm constructors must
be passed argument distributed=True, method .compute() should be passed the right
subset of the data for each node, and after the distributed computations are finalized,
function daal4py.daalfini must be called before accessing the results object.
Example:
distributed_qr.pyimport daal4py
import numpy as np
NUM_NODES = daal4py.num_procs() # this the MPI world size
THIS_NODE = daal4py.my_procid() # this is the MPI rank
rng = np.random.RandomState(seed=123)
X_full = rng.standard_normal(size=(100,5))
subsets = np.split(np.arange(100), NUM_NODES)
X_node = X_full[ subsets[THIS_NODE] ]
qr_algo = daal4py.qr(distributed=True)
qr_result = qr_algo.compute(X_node)
daal4py.daalfini() # call before accessing the results
# Matrix R (shape=[ncols, ncols]) is common for all nodes
np.testing.assert_almost_equal(
np.abs( qr_result.matrixR ),
np.abs( np.linalg.qr(X_full).R ),
)
# Matrix Q (size=[nrows, ncols]) will be a subset of the full
# result corresponding to the data from the node only
np.testing.assert_almost_equal(
np.abs( qr_result.matrixQ ),
np.abs( np.linalg.qr(X_full).Q[ subsets[THIS_NODE] ] ),
)
Then execute as follows - example can be executed on a single machine after installing package impi_rt:
mpirun -n 2 python distributed_qr.py
(can also use mpiexec on Linux)
Note
QR factorization, unlike other linear algebra procedures, does not have a strictly unique solution - if the signs (+/-) of numbers are flipped for a particular column in both the Q and R matrices, they would still be valid and equivalent QR factorizations of the same original matrix ‘X’.
Procedures like Cholesky decomposition are typically constrained to have only positive signs in the main diagonal in order to make the results deterministic, but this is not always the case for QR in most software, hence the example above takes the absolute values when comparing results from different libraries.
In this simple example, all of the data was generated on each node and then subdivided; and then the result was broadcasted to all nodes, but in practice:
One might want to collect and serialize the result on only one node, which could be done for example by adding a condition like
if THIS_NODE == 0. For the particular case of QR, oftentimes only the R matrix is of interest, so it can be saved from only one of the nodes.One might have different files with different names for each node. Likely, one might want to have logic in the code to load different subsets of the data based on the rank of the process, for example
pl.read_parquet(f"file{daal4py.my_procid()}.parquet").
Note that the example above used functions from daal4py to get the world size
(daal4py.num_procs) and process rank (daal4py.my_procid) from MPI. Module daal4py
provides simple wrappers over these two MPI functions only, but for further MPI functionalities,
one can use the package mpi4py together with daal4py.
Same example calling MPI functionalities from mpi4py instead:
distributed_qr_mpi4py.pyimport daal4py
import numpy as np
from mpi4py import MPI
comm = MPI.COMM_WORLD
NUM_NODES = comm.Get_size()
THIS_NODE = comm.Get_rank()
rng = np.random.RandomState(seed=123)
X_full = rng.standard_normal(size=(100,5))
subsets = np.split(np.arange(100), NUM_NODES)
X_node = X_full[ subsets[THIS_NODE] ]
qr_algo = daal4py.qr(distributed=True)
qr_result = qr_algo.compute(X_node)
daal4py.daalfini() # call before accessing results
# Matrix R (shape=[ncols, ncols]) is common for all nodes
np.testing.assert_almost_equal(
np.abs( qr_result.matrixR ),
np.abs( np.linalg.qr(X_full).R ),
)
# Matrix Q (size=[nrows, ncols]) will be a subset of the full
# result corresponding to the data from the node only
np.testing.assert_almost_equal(
np.abs( qr_result.matrixQ ),
np.abs( np.linalg.qr(X_full).Q[ subsets[THIS_NODE] ] ),
)
Can be executed the same way as before:
mpirun -n 2 python distributed_qr_mpi4py.py
Supported algorithms
The following algorithms in daal4py have support for distributed mode: